High resolution spatial analysis of the whole transcriptome is now here with Visium HD! See up to three orders of magnitude improvement in resolution compared with the previous generation of Visium.
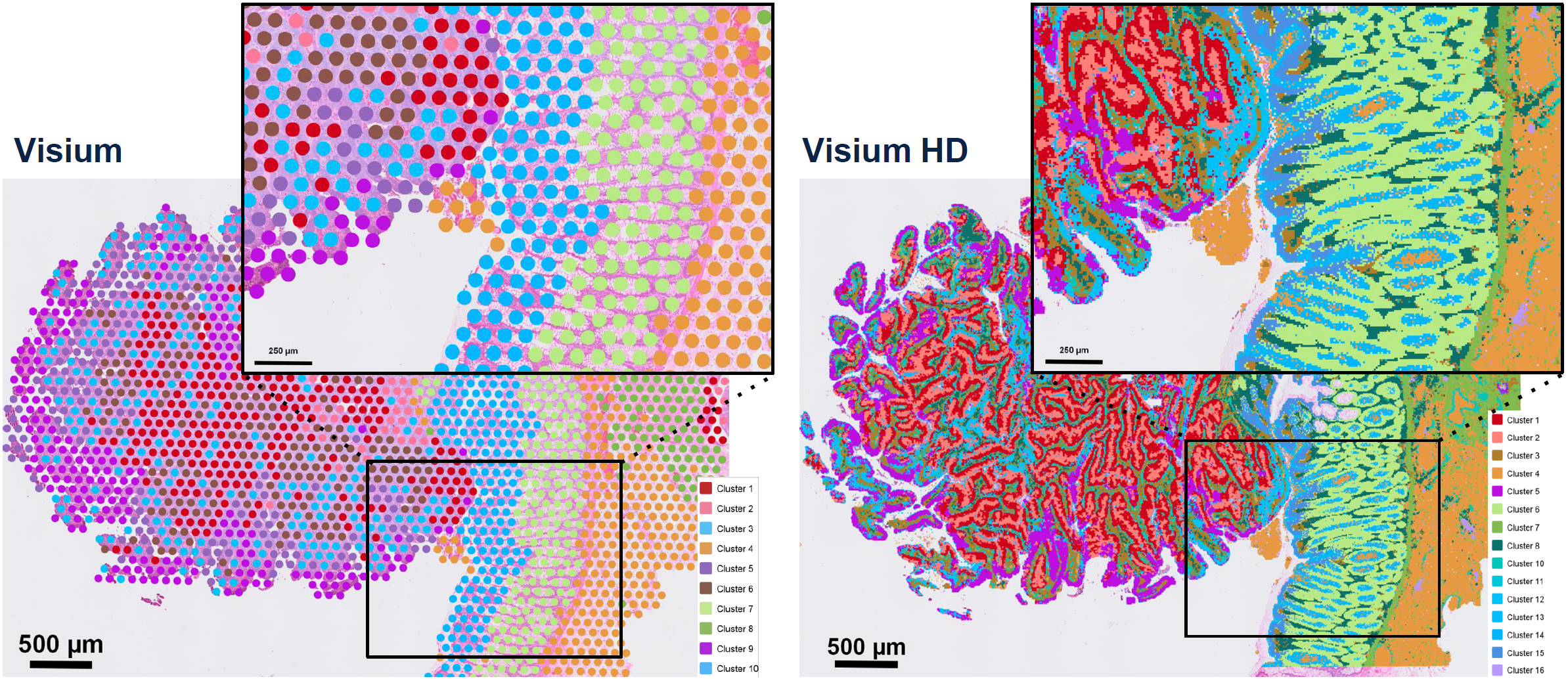
Human colorectal cancer analyzed with Visium and Visium HD assays (courtesy of 10x Genomics).
Summary of features comparing Visium and Visium HD:
| Features | Visium (standard) | Visium HD |
|---|---|---|
| Distribution of capture oligos | 55 micron spots | 2 micron tiles |
| Number of distinct capture elements | ~5,000 | ~10 million |
| Whole transcriptome detection | >18,000 (human) or >20,000 (mouse) gene probe sets | >18,000 (human) or >20,000 (mouse) gene probe sets |
| Sample capture area | 6.5 x 6.5 mm | 6.5 x 6.5 mm |
| Capture from standard FFPE slides | Yes | Yes |
The new spatial assay from 10x Genomics probes mRNAs from FFPE tissue sections and captures those probes onto the Visium HD slide. Each Visium HD slide has the same 6.5 x 6.5mm capture area as previous Visium products but is covered with about 10 million uniquely-barcoded oligonucleotide squares. These 2 micron tiles are arrayed in a continuous lawn across the entire capture area. The barcoded oligos capture and facilitate tagging of hybridized probes, permitting determination of the specific transcriptome of each sector at cellular-level resolution (note that 10x Genomics recommends binning adjacent tiles to 8 x 8 microns for initial analysis). Expression profiles can then be compared and mapped back to the tissue morphology by overlay on a standard H&E stained or immunofluorescence image of the tissue slice.
The Arts & Sciences Imaging Center is a participant in the 10x Genomics Visium Enablement Program, which verifies 10x Genomics authorized reagents and methods for spatial transcriptomics. Contact us to learn more or to discuss your project.
In case you missed it, you can view the seminar "The latest and greatest: new developments in single cell and spatial transcriptomics from 10x Genomics" the we hosted on campus in June 2024.
(Images provided by 10x Genomics)